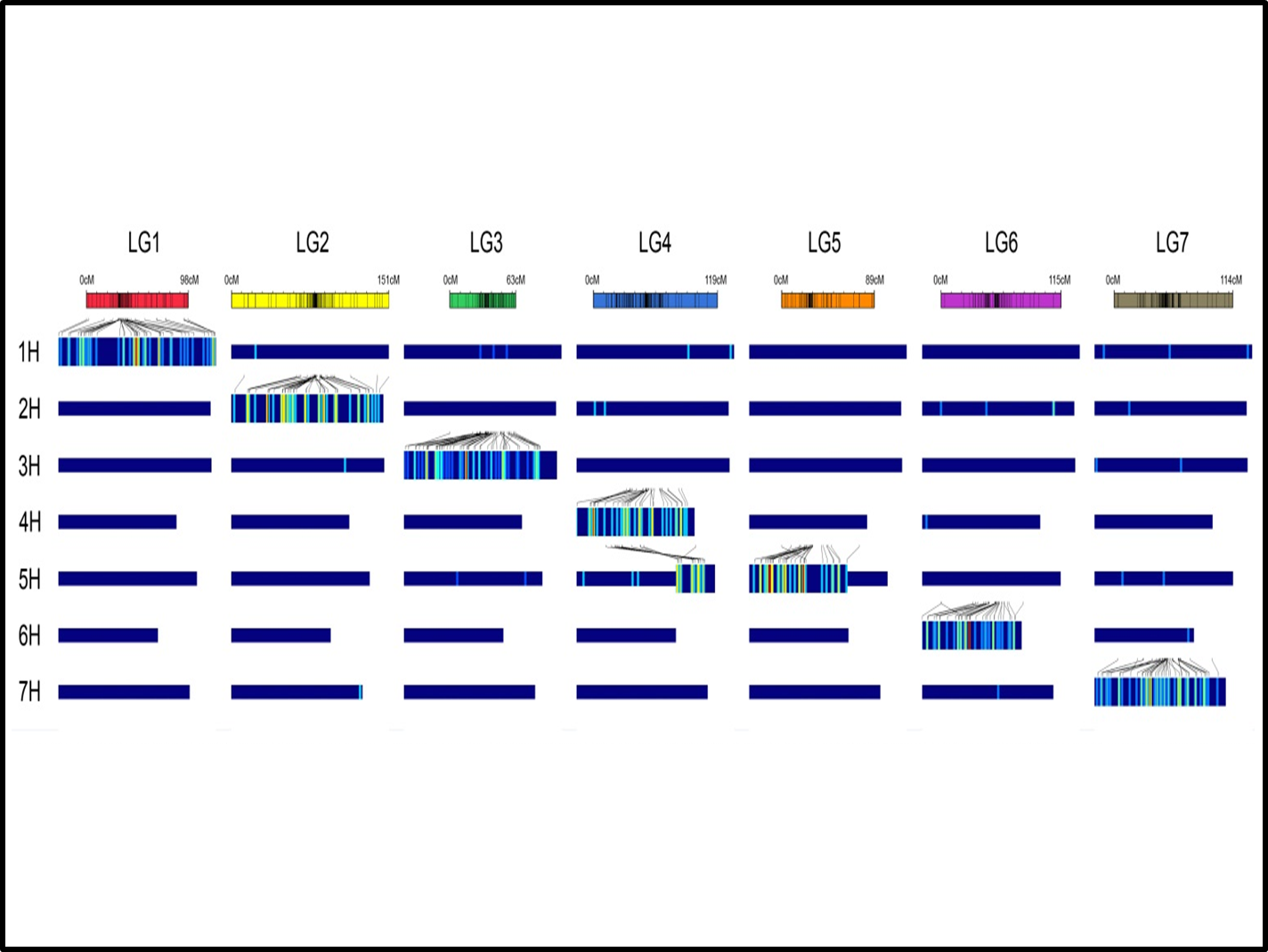
The perennial ryegrass GenomeZipper
Contact person: Bruno Studer
Partners: Matthias Pfeifer, Mihaela Martis, Torben Asp, Klaus F. X. Mayer, Thomas Lübberstedt, Stephen Byrne and Ursula Frei
Project description: Whole-genome sequences established for model and major crop species constitute a key resource for advanced genomic research. For outbreeding forage and turf grass species like ryegrasses (Lolium spp.), such resources are yet to be developed.
Here, we present a model of the perennial ryegrass (Lolium perenne L.) genome on the basis of conserved synteny to barley (Hordeum vulgare L.) and the model grass genome Brachypodium (Brachypodium distachyon L.), as well as rice (Oryza sativa L.) and sorghum [Sorghum bicolor (L.) Moench] (Figure 1). A transcriptome-based genetic linkage map of perennial ryegrass served as a scaffold to establish the chromosomal arrangement of syntenic genes from model grass species. This scaffold revealed a high degree of synteny and macro-collinearity, and was then utilised to anchor a collection of perennial ryegrass genes in silico to their predicted genome position. This resulted in the unambiguous assignment of 3,315 out of 8,876 previously unmapped genes to the respective chromosomes. In total, the GenomeZipper incorporates 4,035 conserved grass gene loci which were used for the first genome-wide sequence divergence analysis between perennial ryegrass, barley, Brachypodium, rice, and sorghum.
The perennial ryegrass GenomeZipper is an ordered, information-rich genome scaffold, facilitating map-based cloning and genome assembly in perennial ryegrass and closely related Poaceae species. It also represents a milestone in describing synteny between perennial ryegrass and fully sequenced model grass genomes, thereby increasing our understanding of genome organization and evolution in the most important temperate forage and turf grass species.
The perennial ryegrass GenomeZipper including GenBank accession numbers and detailed marker information is available online under: external page MIPS Perennial Ryegrass Database