A transcriptome map of perennial ryegrass
Contact person: Bruno Studer
Partners: Stephen Byrne, Rasmus O. Nielsen, Frank Panitz, Christian Bendixen, Md. Shofiqul Islam, Matthias Pfeifer, Thomas Lübberstedt and Torben Asp
Project description: Single Nucleotide Polymorphisms (SNPs) are increasingly becoming the DNA marker system of choice due to their prevalence in the genome and their ability to be used in highly multiplexed genotyping assays. Although needed in high numbers for genome-wide marker profiles and genomics-assisted breeding, a surprisingly low number of validated and mapped SNPs are currently available for perennial ryegrass.
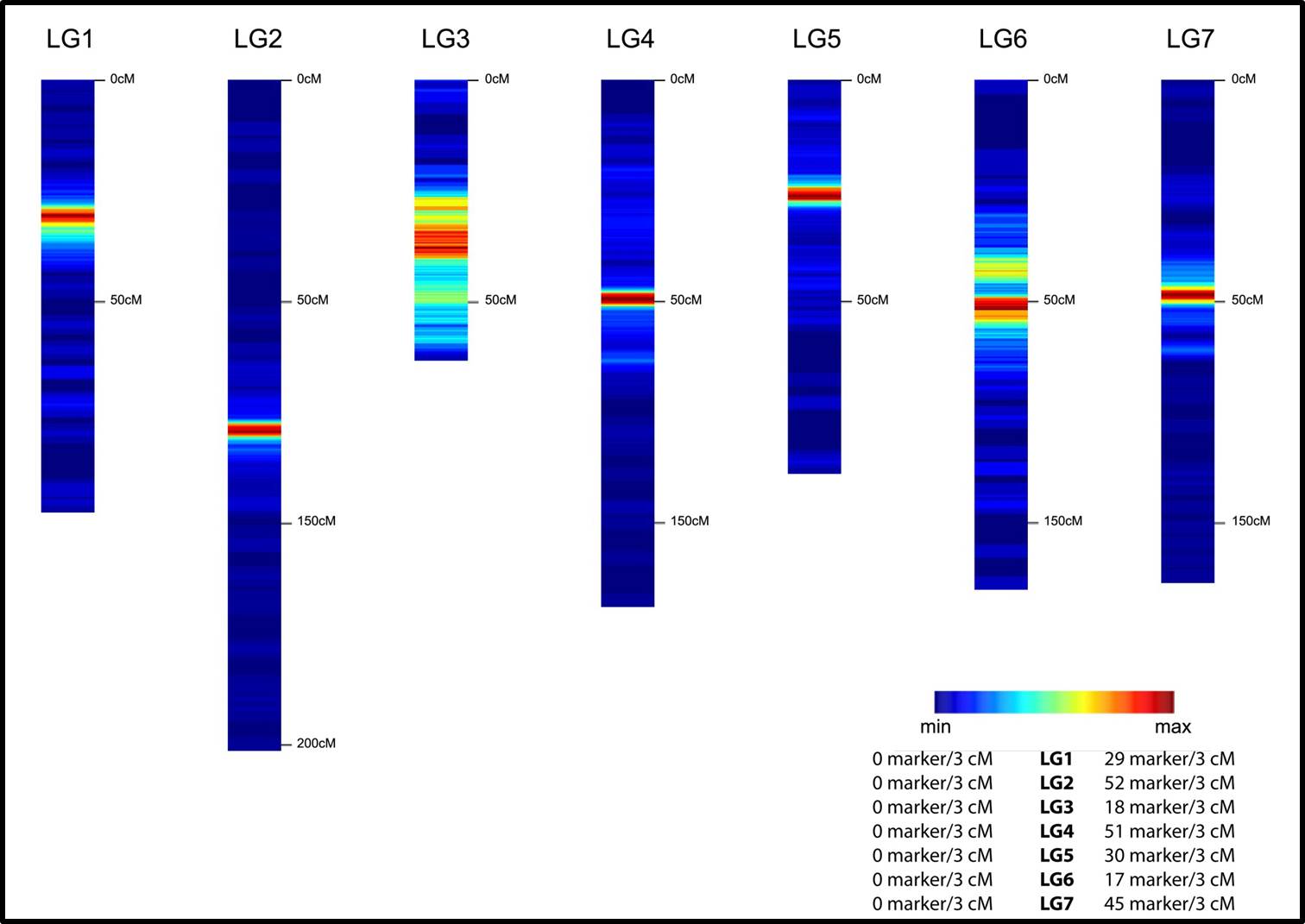
A perennial ryegrass unigene set representing 9,399 genes was used as a reference for the assembly of 802,156 high quality reads generated by 454 transcriptome sequencing and for in silico SNP discovery. Out of more than 15,433 SNPs in 1,778 unigenes fulfilling highly stringent assembly and detection parameters, a total of 768 SNP markers were selected for GoldenGate genotyping in 184 individuals of the perennial ryegrass mapping population VrnA, a population being previously evaluated for important agronomic traits. A total of 592 (77%) of the SNPs tested were successfully called with a cluster separation above 0.9. Of these, 509 (86%) genic SNP markers segregated in the VrnA mapping population, out of which 495 were assigned to map positions. The genetic linkage map presented here comprises a total of 838 DNA markers (767 gene-derived markers) and spans 750 centi Mogan (cM) with an average marker interval distance of less than 0.9 cM. Moreover, it locates 732 expressed genes involved in a broad range of molecular functions of different biological processes in the perennial ryegrass genome.
Here, we present an efficient approach of using next generation sequencing data for SNP discovery, and the successful design of a 768-plex Illumina GoldenGate genotyping assay in a complex genome. The ryegrass SNPs along with the corresponding transcribed sequences represent a milestone in the establishment of genetic and genomics resources available for this species and constitute a further step towards molecular breeding strategies. Moreover, the high density genetic linkage map predominantly based on gene-associated DNA markers provides an important tool for the assignment of candidate genes to QTL, functional genomics and the integration of genetic and physical maps in perennial ryegrass, one of the most important temperate grassland species.