Kyuss v2.0 – THE reference genome sequence for perennial ryegrass
Ultra-long ONT reads with Hi-C enabled the generation of a nearly perfect genome assembly of Kyuss, a doubled haploid perennial ryegrass genotype.
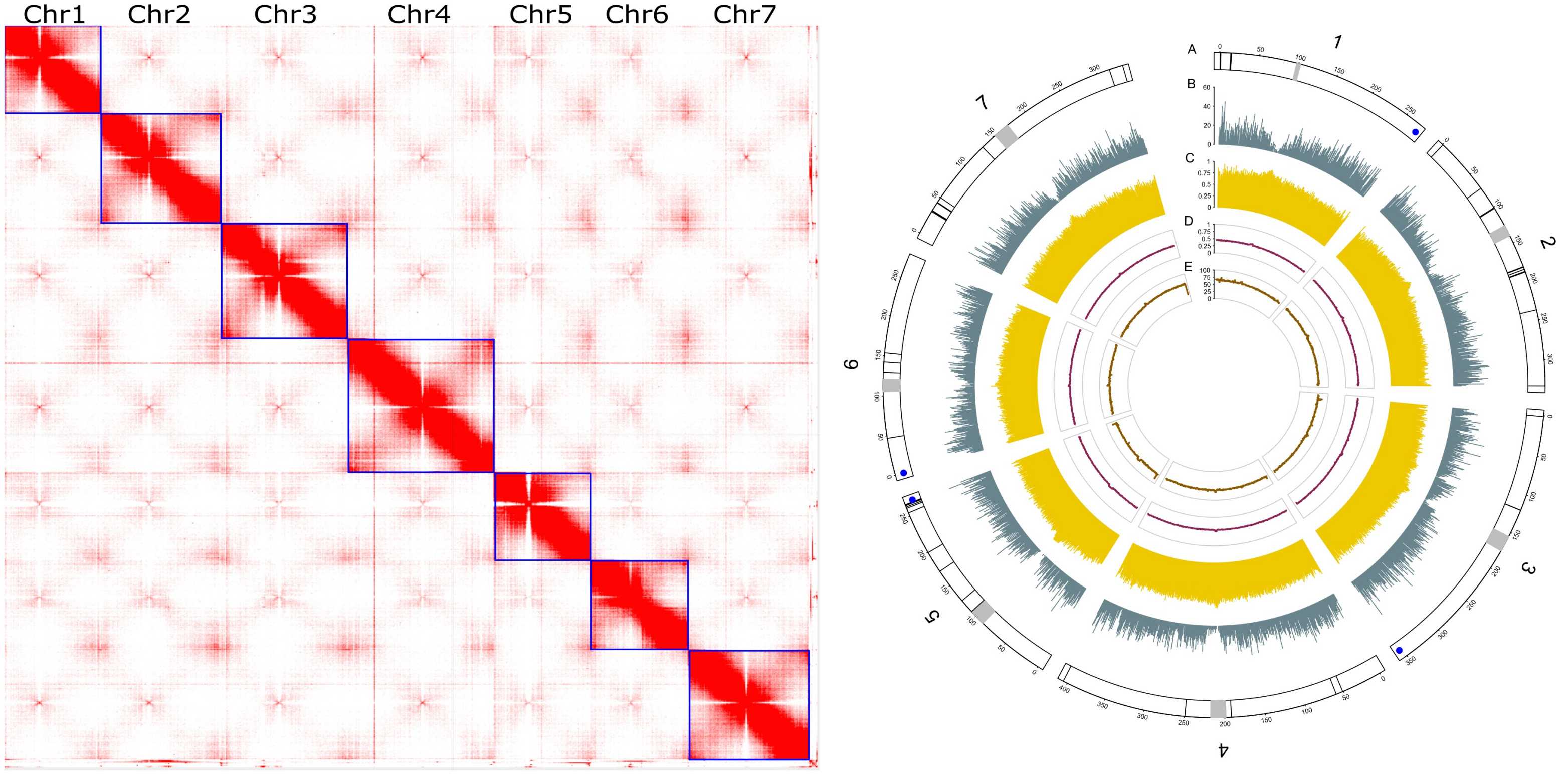
Perennial ryegrass (Lolium perenne L.) is one of the most important forage and turf grass species, widely grown in temperate regions across the world. However, genomic resources, especially reference-level genome assemblies, are scarce for this species. Previously, Kyuss, a doubled haploid perennial ryegrass genotype was sequenced with ultra-long Oxford Nanopore Technologies (ONT) and a chromosome-level genome assembly (Kyuss v1.0) was generated with contigs anchored to pseudo-chromosomes based on genome synteny to barley (Hordeum vulgare L.). As perennial ryegrass and barley diverged around 30 million years ago, structural variations between the genome of those two species may have led to scaffolding errors in the Kyuss v1.0 assembly.
To generate an improved assembly without scaffolding errors for Kyuss, we re-assembled Kyuss with the same ultra-long ONT data but added newly generated chromosome conformation capture (Hi-C) data for scaffolding and pseudo-chromosome construction. The resulting chromosome-level assembly (Kyuss v2.0) is of highest quality with contig N50 of 120 Mb, scaffold N50 of 333 Mb, 99% completeness, 99.999% base-level accuracy as well as accurate sequence order and orientation.
Kyuss v2.0 will serve as THE reference genome sequence for perennial ryegrass and other closely related species and will facilitate genomic applications such as genome-wide association studies and genomic selection to advance forage and turf grass research and breeding.
Citation:
Yutang C, Kölliker R, Mascher M, Copetti D, Himmelbach A, Stein N and Studer B (2024) An improved chromosome-level genome assembly of perennial ryegrass (Lolium perenne L.). Gigabyte, 2024, 1-11.
external page https://doi.org/10.46471/gigabyte.112