High genomic plasticity and unique features of Xanthomonas translucens pv. graminis
Complete genome sequences revealed a high level of chromosomal rearrangements and to identify features potentially involved in pathogenicity.
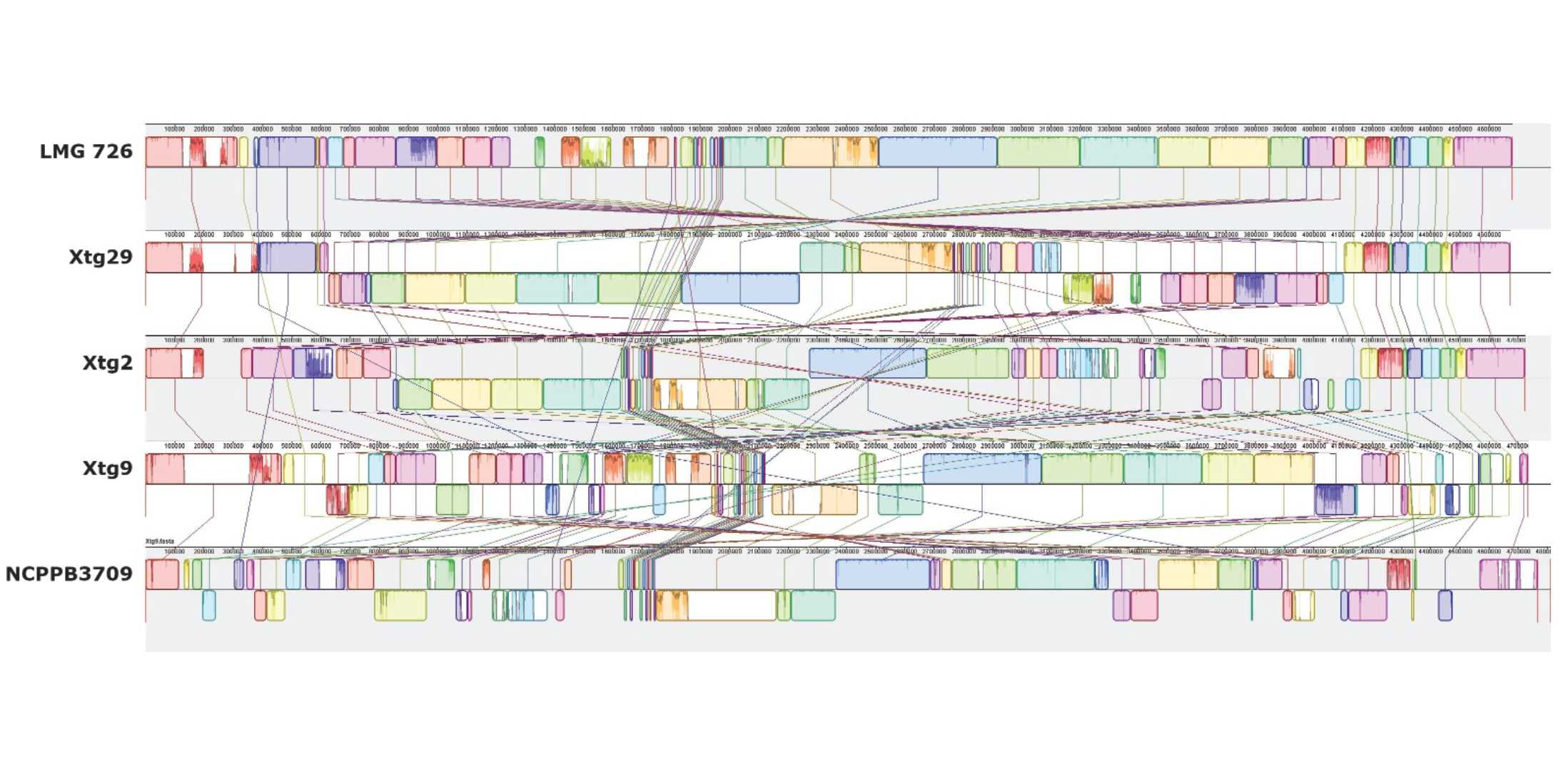
Xanthomonas translucens pv. graminis (Xtg) is a bacterial pathogen that can cause serious damage to forage grasses. The disease can occur in a wide range of grass species, but mainly affects Italian ryegrass (Lolium multiflorum Lam.), an important component of hay and silage production. To efficiently breed for Xtg resistance, it is crucial to understand the genetic composition of the pathogen and the mechanisms used to infect the plant.
So far, mostly draft genome sequences were available for this pathovar, preventing comprehensive genomic analyses. We have sequenced four additional strains of Xtg with high-coverage long read sequencing technology and assembled high-quality, complete genomes for all four strains. Together with the currently available complete genome sequences for other pathovars of the species, these new genome sequences allowed for an in-depth comparative analysis focused on Xtg.
All Xtg genome sequences showed a high level of rearrangements between strains. Additionally, a high number of transposable elements were found to be exclusive to Xtg, with 413 to 457 insertion/excision elements per strain. Such mobile elements can facilitate gene duplications, chromosomal rearrangements, horizontal gene transfer, and may cause the loss of gene function. They are thus likely involved in the observed high genomic plasticity and could play a key role in the high adaptability of Xtg.
The pathovar was found to lack many features usually associated with pathogenicity in Xanthomonas spp., such as the type IV secretion system, or transcription activator-like effectors (TALEs). It also possessed the smallest set of type III effectors, and a highly divergent type IV pilus compared to the rest of the species. This loss or divergence of main virulence features in Xtg may be due to its high genomic plasticity and may be associated with its ability to infect a large range of plant hosts, by allowing the bacteria to evade plant defense mechanisms.
In total, 1,409 genes were found to be exclusive to Xtg, with 536 of them being shared by all strains. Excluding transposable elements, a total of 148 annotated genes were found to be exclusive to Xtg and shared by all strains. These included genes that are known to play a role in pathogenicity, such as a filamentous hemagglutinin, a YadA family autotransporter adhesin, ten TonB-dependent receptors, and eleven plant cell wall degradative enzymes. Furthermore, three XopE and XopX family effectors were found in all Xtg strains, while the other pathovars possessed two or less. These constitute a set of potential virulence factors and investigating these genes further will allow to better understand the virulence and host range of Xtg.
These results provide valuable insights into potential adaptation mechanisms found in Xtg, and the identified virulence features will help to better understand the biology of Xtg, laying the foundations for the development of new resistant forage grass cultivars.
This research was supported by the Swiss National Science Foundation (Grant No: IZCOZO_177062) and was based upon work from COST Action CA16107 EuroXanth, supported by COST (European Cooperation in Science and Technology).
Citation:
Goettelmann F, Koebnik R, Roman-Reyna V, Studer B and Kölliker R (2023) High genomic plasticity and unique features of Xanthomonas translucens pv. graminis revealed through comparative analysis of complete genome sequences. BMC Genomics 24:741.
external page https://doi.org/10.1186/s12864-023-09855-8